[1]:
import jax
import jax.numpy as jnp
from jax import random, vmap, jit
import numpy as np
import pymc as pm
import pymc.sampling_jax
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
from sgmcmcjax.samplers import build_sgld_sampler, build_sgldCV_sampler
import optax
from sgmcmcjax.optimizer import build_optax_optimizer
print(f"Running on PyMC v{pm.__version__}")
/Users/jeremiecoullon/Documents/research/jax/sgmcmc_samplers/venv/lib/python3.8/site-packages/aesara/link/jax/dispatch.py:86: UserWarning: JAX omnistaging couldn't be disabled: Disabling of omnistaging is no longer supported in JAX version 0.2.12 and higher: see https://github.com/google/jax/blob/main/design_notes/omnistaging.md.
warnings.warn(f"JAX omnistaging couldn't be disabled: {e}")
/Users/jeremiecoullon/Documents/research/jax/sgmcmc_samplers/venv/lib/python3.8/site-packages/pymc/sampling_jax.py:34: UserWarning: This module is experimental.
warnings.warn("This module is experimental.")
Running on PyMC v4.0.0b2
Sample from a PyMC model using SGMCMCJax¶
todo: - calculate ESS/sec and print out for sgld, sgld-cv, and NUTS - try another model from the examples: https://docs.pymc.io/en/v3/nb_examples/index.html ? A lot of these don’t use V4 (example PMF would be good to try out as it takes an hour for NUTS)
[2]:
from pymc.sampling_jax import get_jaxified_graph
def build_pymc_loglik_logprior(model):
original_data = []
dummy_data_inputs = []
for observed_RV in model.observed_RVs:
data = model.rvs_to_values[observed_RV]
dummy_data_input = data.type()
# TODO: You should revert these inplace changes after you're done
model.rvs_to_values[observed_RV] = dummy_data_input
original_data.append(data.data)
dummy_data_inputs.append(dummy_data_input)
# build loglik
loglike_fn = get_jaxified_graph(
inputs=model.value_vars + dummy_data_inputs,
outputs=[model.datalogpt],
)
def pymc_loglikelihood(theta, x):
return loglike_fn(*theta, x)[0]
# build logprior
logp_fn = get_jaxified_graph(
outputs=[model.varlogpt],
)
def pymc_logprior(theta):
return logp_fn(*theta)[0]
return pymc_loglikelihood, pymc_logprior
PyMC¶
[4]:
# generate dataset
N, D = 10_000, 10
key = random.PRNGKey(0)
X_data = random.normal(key, shape=(N, D))
with pm.Model() as model:
x = pm.Normal("x", 0, 10, shape=D)
y = pm.Normal("y", x, observed=X_data)
model
[4]:
$$
\begin{array}{rcl}
\text{x} &\sim & \operatorname{N}(0,~10)\\\text{y} &\sim & \operatorname{N}(\text{x},~1)
\end{array}
$$
run PyMC sampler¶
[5]:
%%time
with model:
posterior = pm.sample(10_000, chains=1)
# adding this line adds a lot of time.
# is there a "block_until_ready()" missing somewhere?..
print(posterior.posterior.x.values[0,:][-1])
Auto-assigning NUTS sampler...
INFO:pymc:Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
INFO:pymc:Initializing NUTS using jitter+adapt_diag...
Sequential sampling (1 chains in 1 job)
INFO:pymc:Sequential sampling (1 chains in 1 job)
NUTS: [x]
INFO:pymc:NUTS: [x]
100.00% [11000/11000 00:30<00:00 Sampling chain 0, 0 divergences]
Sampling 1 chain for 1_000 tune and 10_000 draw iterations (1_000 + 10_000 draws total) took 30 seconds.
INFO:pymc:Sampling 1 chain for 1_000 tune and 10_000 draw iterations (1_000 + 10_000 draws total) took 30 seconds.
Only one chain was sampled, this makes it impossible to run some convergence checks
INFO:pymc:Only one chain was sampled, this makes it impossible to run some convergence checks
[-0.00409768 0.00598126 0.00143343 0.01084321 -0.00564738 0.0039423
0.01297556 -0.02975454 -0.01849593 -0.01484458]
CPU times: user 49.3 s, sys: 59.2 s, total: 1min 48s
Wall time: 2min 56s
run using SGMCMCJax¶
[6]:
pymc_loglikelihood, pymc_logprior = build_pymc_loglik_logprior(model)
[7]:
# build sampler
batch_size = int(0.1*N)
dt = 1e-5
rvs = [rv.name for rv in model.value_vars]
init_position_dict = model.compute_initial_point()
init_position = [init_position_dict[rv] for rv in rvs]
# ========
my_sampler_pymc = build_sgld_sampler(dt,
pymc_loglikelihood,
pymc_logprior,
(X_data,),
batch_size)
# get MAP for SGLD-CV
# Adam
batch_size_adam = int(0.1 * N)
dt_adam = 1e-3
opt = optax.adam(learning_rate=dt_adam)
optimizer = build_optax_optimizer(opt, pymc_loglikelihood, pymc_logprior, (X_data,), batch_size_adam)
Niters = 10_000
opt_params, log_post_list = optimizer(key, Niters, init_position)
# define SGLD-CV
my_sampler_pymc_CV = build_sgldCV_sampler(dt,
pymc_loglikelihood,
pymc_logprior,
(X_data,),
batch_size,
opt_params)
[8]:
%%time
Nsamples = 10_000
# run SGLD
samples_pymc = my_sampler_pymc(key, Nsamples, init_position)
# run SGLD-CV
samples_pymc_CV = my_sampler_pymc_CV(key, Nsamples, init_position)
# result are in a PyTree
_ = samples_pymc[0].block_until_ready()
_ = samples_pymc_CV[0].block_until_ready()
CPU times: user 4.32 s, sys: 78.5 ms, total: 4.4 s
Wall time: 4.43 s
compare posterior samples¶
[9]:
chain_num = 0
idx = 0
plt.rcParams.update({'font.size': 16})
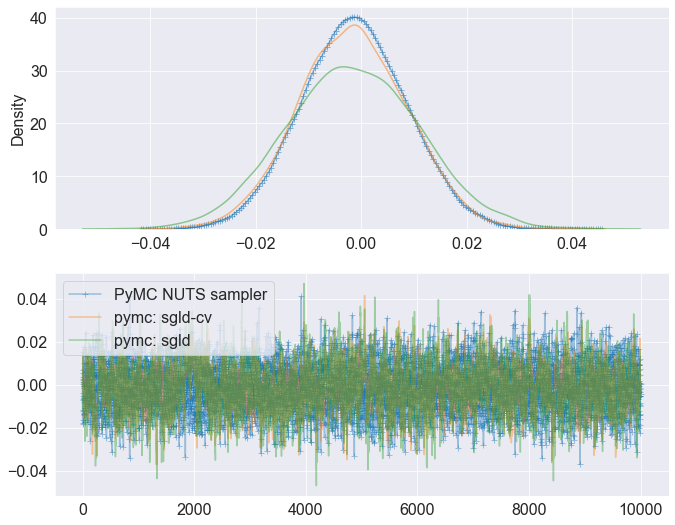
fig, ax = plt.subplots(2, figsize=(11, 9))
sns.kdeplot(posterior.posterior.x.values[chain_num, :, idx],
label="PyMC NUTS sampler", alpha=0.5, ax=ax[0], marker="+")
sns.kdeplot(samples_pymc_CV[0][:,idx], label="pymc: sgld-cv", alpha=0.5, ax=ax[0])
sns.kdeplot(samples_pymc[0][:,idx], label="pymc: sgld", alpha=0.5, ax=ax[0])
ax[1].plot(posterior.posterior.x.values[chain_num, :, idx],
label="PyMC NUTS sampler", alpha=0.4, marker="+")
ax[1].plot(samples_pymc_CV[0][:,idx], label="pymc: sgld-cv", alpha=0.4)
ax[1].plot(samples_pymc[0][:,idx], label="pymc: sgld", alpha=0.4)
plt.legend()
[9]:
<matplotlib.legend.Legend at 0x7fe86dd97d00>
[ ]:
[ ]:
[ ]: