[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import jax.numpy as jnp
import jax
from jax import random, jit
from tqdm.auto import tqdm
from sgmcmcjax.kernels import build_sgld_kernel, build_psgld_kernel, build_sgldAdam_kernel, build_sghmc_kernel
import tensorflow_datasets as tfds
from flax import linen as nn
Flax CNN¶
Colab notebook showing how to run this example on a TPU
[2]:
class CNN(nn.Module):
"""A simple CNN model."""
@nn.compact
def __call__(self, x):
x = nn.Conv(features=32, kernel_size=(3, 3))(x)
x = nn.relu(x)
x = nn.avg_pool(x, window_shape=(2, 2), strides=(2, 2))
x = nn.Conv(features=64, kernel_size=(3, 3))(x)
x = nn.relu(x)
x = nn.avg_pool(x, window_shape=(2, 2), strides=(2, 2))
x = x.reshape((x.shape[0], -1)) # flatten
x = nn.Dense(features=256)(x)
x = nn.relu(x)
x = nn.Dense(features=10)(x)
x = nn.log_softmax(x)
return x
[3]:
cnn = CNN()
def loglikelihood(params, x, y):
x = x[jnp.newaxis] # add an axis so that it works for a single data point
logits = cnn.apply({'params':(params)}, x)
label = jax.nn.one_hot(y, num_classes=10)
return jnp.sum(logits*label)
def logprior(params):
return 1.
@jit
def accuracy_cnn(params, X, y):
target_class = y
predicted_class = jnp.argmax(cnn.apply({'params':(params)}, X), axis=1)
return jnp.mean(predicted_class == target_class)
def get_datasets():
"""Load MNIST train and test datasets into memory."""
ds_builder = tfds.builder('mnist')
ds_builder.download_and_prepare()
train_ds = tfds.as_numpy(ds_builder.as_dataset(split='train', batch_size=-1))
test_ds = tfds.as_numpy(ds_builder.as_dataset(split='test', batch_size=-1))
train_ds['image'] = jnp.float32(train_ds['image']) / 255.
test_ds['image'] = jnp.float32(test_ds['image']) / 255.
return train_ds, test_ds
train_ds, test_ds = get_datasets()
X_train_s = train_ds['image']
y_train_s = jnp.array(train_ds['label'])
X_test_s = test_ds['image']
y_test_s = jnp.array(test_ds['label'])
data = (X_train_s, y_train_s)
batch_size = int(0.01*data[0].shape[0])
WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
[4]:
def run_sgmcmc(key, Nsamples, init_fn, my_kernel, get_params, accuracy_rate=100):
"Run SGMCMC sampler and return the test accuracy list"
accuracy_list = []
params = cnn.init(key, jnp.ones([1,28,28,1]))['params']
key, subkey = random.split(key)
state = init_fn(subkey, params)
for i in tqdm(range(Nsamples)):
key, subkey = random.split(key)
state = my_kernel(i, subkey, state)
if i%accuracy_rate==0:
accuracy_list.append(accuracy_cnn(get_params(state), X_test_s, y_test_s))
return accuracy_list
SGLD¶
[5]:
%%time
init_fn, sgld_kernel, get_params = build_sgld_kernel(5e-6, loglikelihood, logprior, data, batch_size)
sgld_kernel = jit(sgld_kernel)
Nsamples = 2000
accuracy_list_sgld = run_sgmcmc(random.PRNGKey(0), Nsamples, init_fn, sgld_kernel, get_params)
CPU times: user 27min 30s, sys: 6min 57s, total: 34min 27s
Wall time: 8min 6s
SGHMC¶
[6]:
%%time
init_fn, sghmc_kernel, get_params = build_sghmc_kernel(1e-6, 4, loglikelihood,
logprior, data, batch_size, compiled_leapfrog=False)
Nsamples = 500
accuracy_list_sghmc = run_sgmcmc(random.PRNGKey(0), Nsamples, init_fn,
sghmc_kernel, get_params, accuracy_rate=25)
CPU times: user 30min 22s, sys: 7min 39s, total: 38min 1s
Wall time: 9min 49s
pSGLD¶
[15]:
%%time
init_fn, psgld_kernel, get_params = build_psgld_kernel(1e-3, loglikelihood, logprior, data, batch_size)
psgld_kernel = jit(psgld_kernel)
Nsamples = 2000
accuracy_list_psgld = run_sgmcmc(random.PRNGKey(0), Nsamples, init_fn, psgld_kernel, get_params)
CPU times: user 13min 5s, sys: 3min 11s, total: 16min 17s
Wall time: 4min 6s
sgldAdam¶
[8]:
%%time
init_fn, sgldAdam_kernel, get_params = build_sgldAdam_kernel(1e-2, loglikelihood, logprior, data, batch_size)
sgldAdam_kernel = jit(sgldAdam_kernel)
Nsamples = 2000
accuracy_list_sgldAdam = run_sgmcmc(random.PRNGKey(0), Nsamples, init_fn, sgldAdam_kernel, get_params)
CPU times: user 28min 18s, sys: 7min 16s, total: 35min 35s
Wall time: 8min 12s
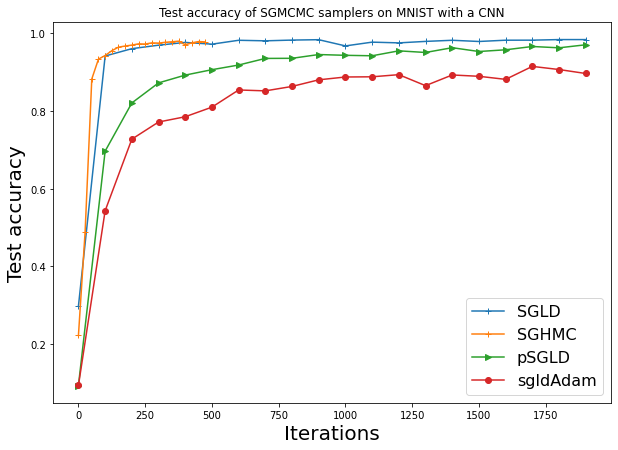
[11]:
plt.figure(figsize=(10,7))
plt.plot(jnp.arange(0,2000,100), accuracy_list_sgld, label="SGLD", marker="+") # 5e-6
plt.plot(jnp.arange(0,500,25), accuracy_list_sghmc, label="SGHMC", marker="+") # dt=1e-6, L=4
plt.plot(jnp.arange(0,2000,100), accuracy_list_psgld, label="pSGLD", marker=">") # 1e-3
plt.plot(jnp.arange(0,2000,100), accuracy_list_sgldAdam, label="sgldAdam", marker="o") # 1e-2
plt.legend(fontsize=16)
plt.title("Test accuracy of SGMCMC samplers on MNIST with a CNN")
plt.xlabel("Iterations", size=20)
plt.ylabel("Test accuracy", size=20)
print(f"sgld: {100*accuracy_list_sgld[-1]:.1f}%")
print(f"sghmc: {100*accuracy_list_sghmc[-1]:.1f}%")
print(f"psgld: {100*accuracy_list_psgld[-1]:.1f}%")
print(f"sgldAdam: {100*accuracy_list_sgldAdam[-1]:.1f}%")
sgld: 98.4%
sghmc: 97.8%
psgld: 97.0%
sgldAdam: 89.6%